PRESENTATION

The Leica TCS SPE is a high-resolution spectral confocal system, based on a Leica DMI4000B microscope.
Location: 2.1.15;
IP: 10.101.134.35 (why?)
Check “What are the costs” at the Users section
| Pos | Mag | NA | IM | Corr. | WD | Glass | Color Code | Pixel size | Step size |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 4x | 0.1 | Air | HCX 4x W | > 1mm | 0.17 mm (#1.5) | >100 um | ||
| 2 | 10x | 0.3 | Air | HC PL APO CS | 1500 um | 0.17 mm (#1.5) | 1.615 um/px | ~8.0 um | |
| 3 | 20x | 0.7 | Air | HC PL APO CS | 590 um | 0.17 mm (#1.5) | 0.750 um/px | ~2.0 um | |
| 4 | option | ||||||||
| 5 | 40x | 1.15 | Oil | ACS APO | 240 um | 0.17 mm (#1.5) | 0.380 um/px | ~1.0 um | |
| 6 | 63x | 1.4 - 0.6 | Oil | HCX PL APO | 100 um | 0.17 mm (#1.5) | 0.242 um/px | ~0.6 um | |
| Option | 2.5x | 0.07 | Air | >2 mm | 0.17 mm (#1.5) | ||||
| Option | 40x | 0.7 | Air | HCX PL APO CS | 200 um | 0.17 mm (#1.5) | 0.380 um/px | ~2.0 um | |
| Option | 63x | 1.15 | Water | HCX 63x W | 150 um | 0.15 - 0.19 mm | 0.242 um/px | ~1.0 um | |
| Option | 40x | 1.25 - 0.75 | Oil | HCX PL APO CS | 100 um | 0.17 mm (#1.5) | 0.380 um/px | ~1.0 um |
Pos Position
Mag Magnification
NA Numerical Aperture
IM Immersion Medium
Corr. Correction Type
WD Working Distance
Glass Coverglass
Pixel size @ 1024×1024; 1x zoom
Step Confocal section thickness (1 AU)
DIC prism available for HCX 63x W
| Position | Excitation (nm) | Emission (nm) | Fluorophores | Model |
|---|---|---|---|---|
| 1 | 330-385 | >420 | DAPI, Hoechst | A |
| 2 | 450-490 | >515 | GFP, Alexa488, cy2, FITC | I3 |
| 3 | 515-560 | >590 | mCherry, YFP, cy3, Alexa546, Alexa568, TRITC | N2.1 |
| "4" | 590-650 | 663-738 | Cy5, Alexa647, ToPRO-3, Methyl Green | Y5 |
For eyepieces only!
Transmitted light
Halogen lamp
Fluorescence
Mercury metal halide bulb
Lifetime: 2000 hours
Integrated fast shutter
Laser lines
488 nm, 532 nm and 635 nm
ATTENTION: this system does not have a UV laser so DAPI staining cannot be imaged in confocal mode. Use red DNA dyes instead (eg ToPro3, Methyl Green)
1 PMT spectral (400-700nm) 1 transmitted light detector;

(non-normalised spectra)
X Y Z T Lambda acquisition
1 x Leica DFC365 FX 1.4MP CCD camera 6.45×6.45 um/pixel
Immersion oil (RI 1.5180)
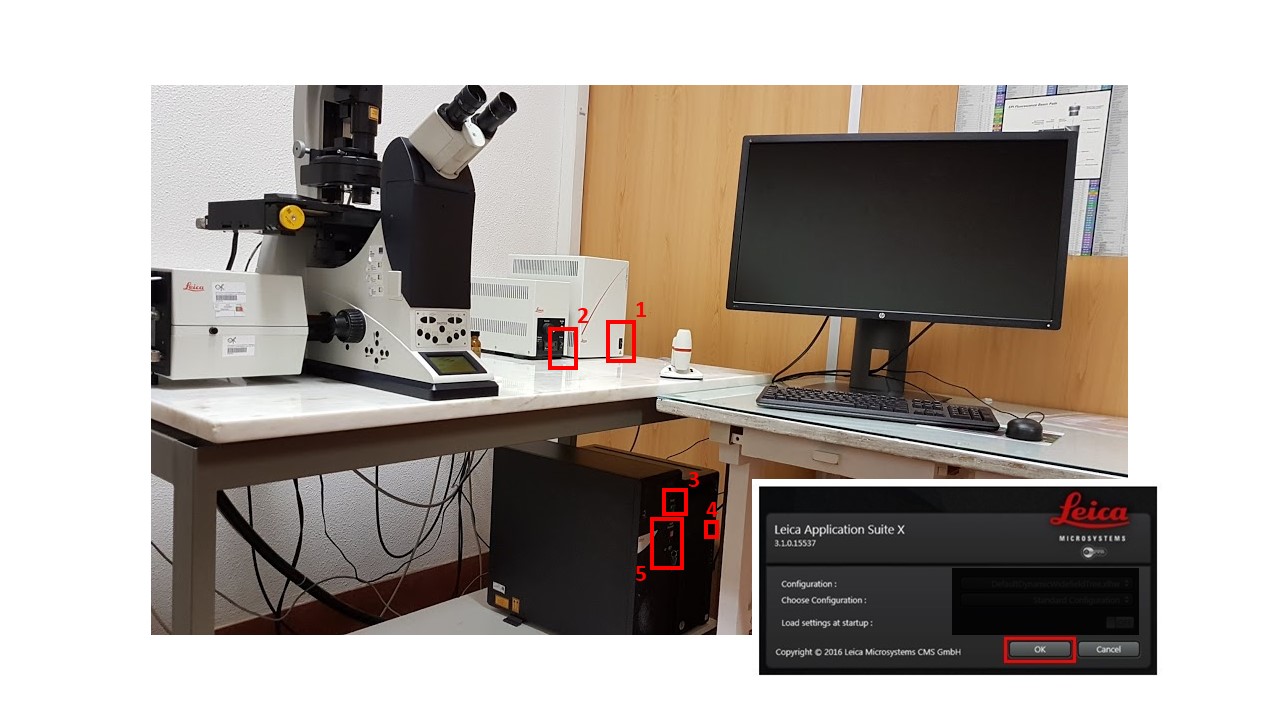
- Check that the objective revolver in the microscope is at its lowest. Turn on microscope control box (1). Allow motorized stage to finish full range of motion for calibration.
- Turn on fluorescence (2), confocal control box (3), and switch on the computer (4). Switch on laser power by turning the (5) key.
- Do not switch fluorescence off for 30 minutes after switching on
- DO NOT switch fluorescence on if the metal casing is still warm, i.e., it was switched off less than 45 minutes before.
- Start LAS X program and click “Ok”.
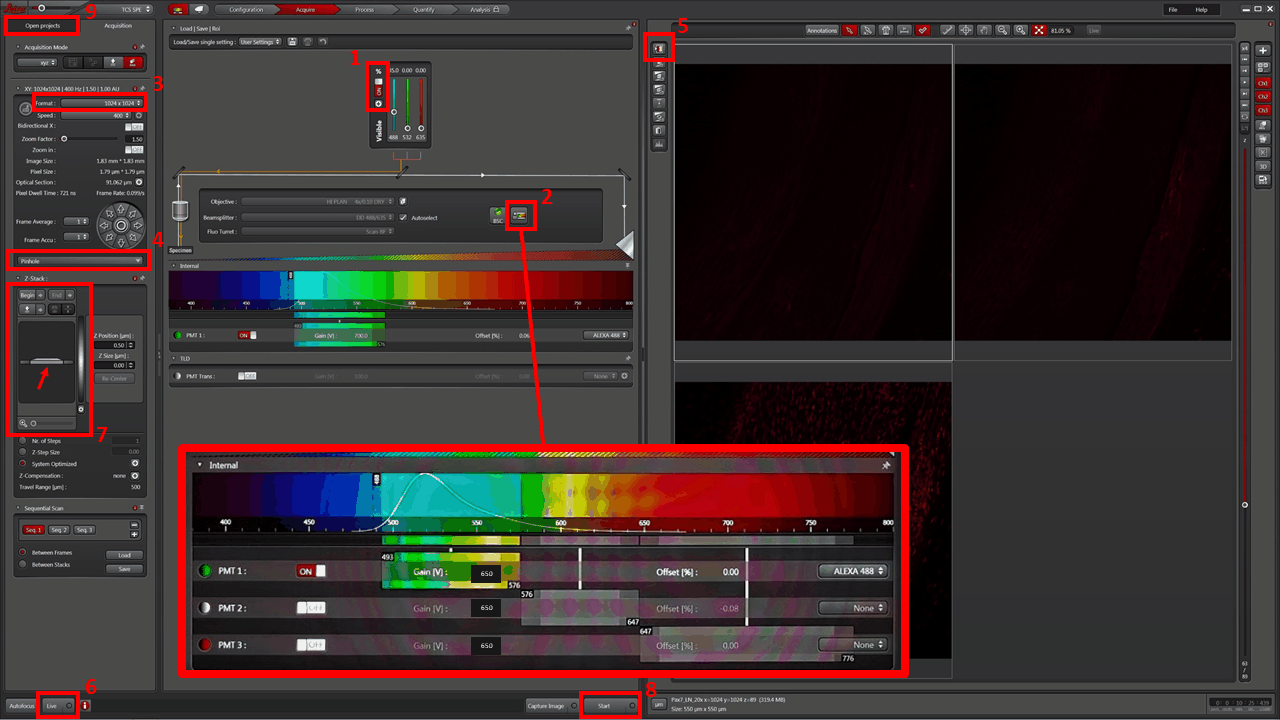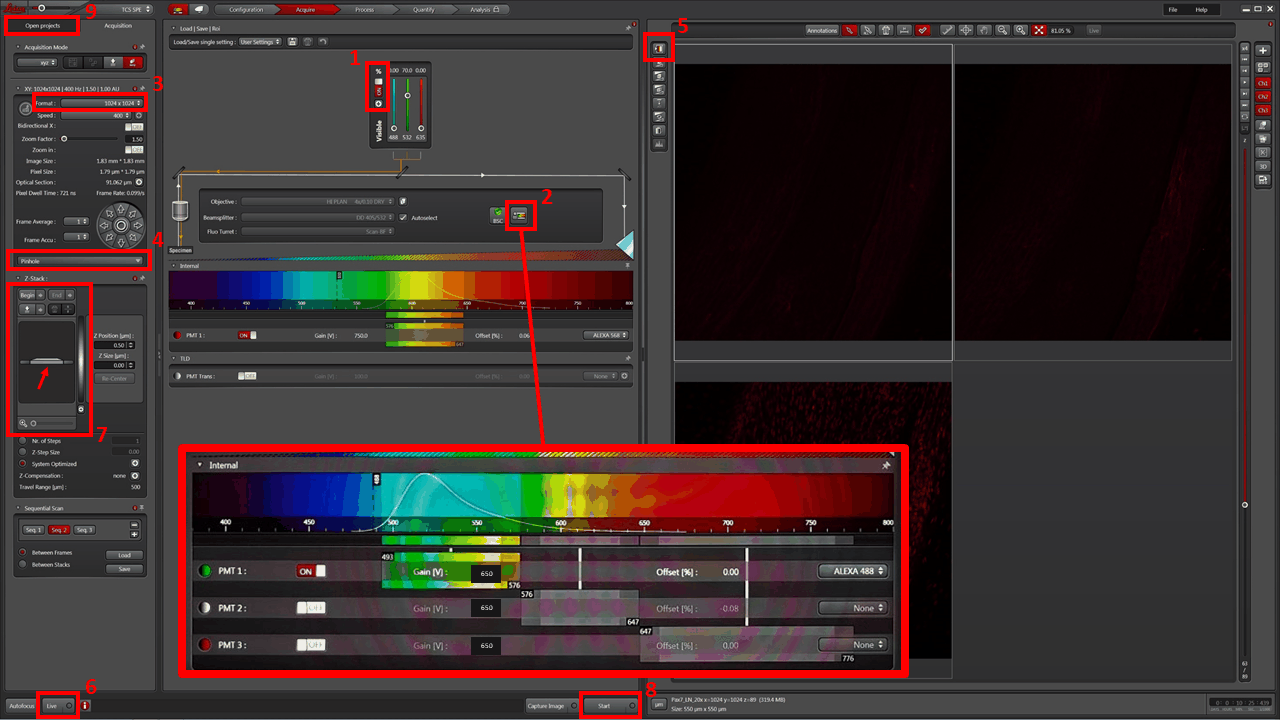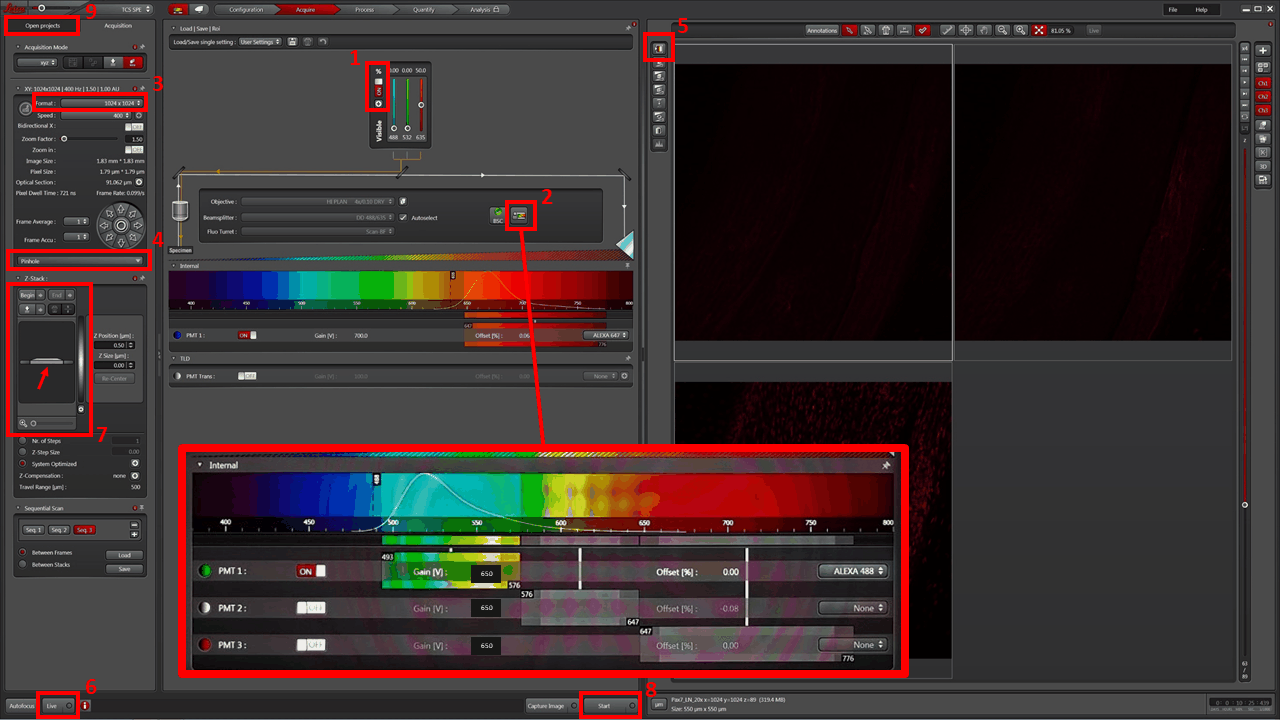
- Press the “+” button (1) and turn on the laser lines you need to use. Unlock them by pressing the “%” On/Off switch.
- Use Dye Assistant wizard (2) to setup your laser path and detector settings. Pick through the fluorophores you wish to use (maximum 3), and assign the desired color lookup tables to each one. Usually, Alexa488 (green), Alexa568 (red), Alexa647 (blue), for example. Press “Apply”.
- You will have set up a SEQUENTIAL image acquisition, with three channels/sequences (Sequential Scan: Seq 1 | Seq 2 | Seq 3). Adjust detector wavelength window FOR EACH SEQUENCE. Good starting values are shown in the (2) blowup:
- Alexa488: 493 – 576
- Alexa568: 576 – 647
- Alexa647: 647 – 776
- Pick image format (3). 1024×1024 is a good compromise for speed/quality, at 400Hz scanning.
- To ensure proper confocal / optical slice acquisition, keep Pinhole (4) at Airy 1.
- Cycle through viewing lookup tables (5) and pick the “saturation” one. Grey levels will appear as reddish-brown, black pixels will show in green and white pixels will show in blue. This will help you choose proper laser power and gain for each channel.
- Select a low magnification objective – 10x is a good starting point. Press the “Eyepiece” selector button on the microscope console, one of the fluorescence cube buttons, and then the “Shutter” button. Look through the eyepieces, move through your slide with the X-Y joystick, and bring your sample into focus with the micrometric knob.
- Click “Live” (6) to preview the confocal image.
- Adjust laser power and gain for each sequence, so that you get an image with very little saturated (blue) pixels. Start at 650 gain and increase to no more than 850 – image will become very noisy. Increasing laser power (start at 20%) will give more signal, but will bleach your sample faster.
- Adjust offset for each sequence so that you see a black background with only a few black/green pixels.
- If you wish to acquire a single optical slice, press “Start” (8) to begin acquisition.
- If you wish to do a stack, click on the light grey bar (arrow in 7) and then use the mouse wheel to travel through your sample in z. Go “up” and set your desired Begin point, and go “down” and set your desired End point. If using the immersion objectives, take care not to hit the slide with the objective (the image will typically become “frozen” in place). The system is very sensitive to pressure on the fast stage, and WILL CRASH DURING A STACK.
- To see your whole programed stack, use the zoom slider underneath the stack window. DO NOT USE the small dark grey bars on the side!! These quickly move the stage and may cause you to hit the slide.
- Pick number of acquisition steps in you stack. For optimal Z-resolution (i.e. if you wish to do 3D reconstruction), choose System Optimized.
- Press “Start” (8) to begin acquisition. If you change objective, readjust settings.
- Your acquisitions will be gathered as *.lif files, in the “Open projects” tab (9). Save it in your image folder. You may create “collections” inside *.lif files by right-clicking on them. A typical structure may be [Project: Experiment name or Date] \ [Collection: slide name or number] \ [Image: name of experimental condition]
- When your work is done, check if anyone is using the microscope after you. If not, press the “%” On/Off to OFF, and click on the “+” button (1) to turn off the lasers. Exit LAS program.
- Turn the laser power key to off, and switch off the lasers control box (green switch).
- Switch off the microscope control box, and the fluorescence.
- Cover the microscope with the plastic cover, taking care not to force any cabling. DO NOT cover the microscope without turning off the microscope control box – fire hazard!
- Transfer your files through Voxel 6 and turn off the computer.
This microscope allows xz / yz imaging and stage experiments (mosaic and mark & find with stacks). Contact Luís for further help with these settings.
To open the images you can either use the Leica LAS X Core software (Windows only!) or FIJI with the latest Bio-Formats plugin installed
LINKS / USER MANUALS
PEOPLE RESPONSIBLE FOR THE EQUIPMENT
 |
 |
|
Telmo Nunes FCUL Microscopy technician mevarrimento@fc.ul.pt Room 2.1.15 |
Luís Marques FCUL Microscopy technician lfmarques@fc.ul.pt Room 8.1.79 / 2.1.15 |
